|
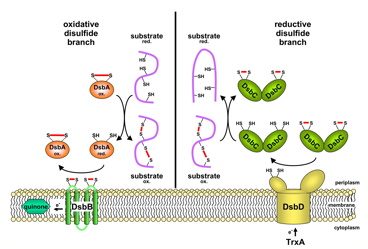
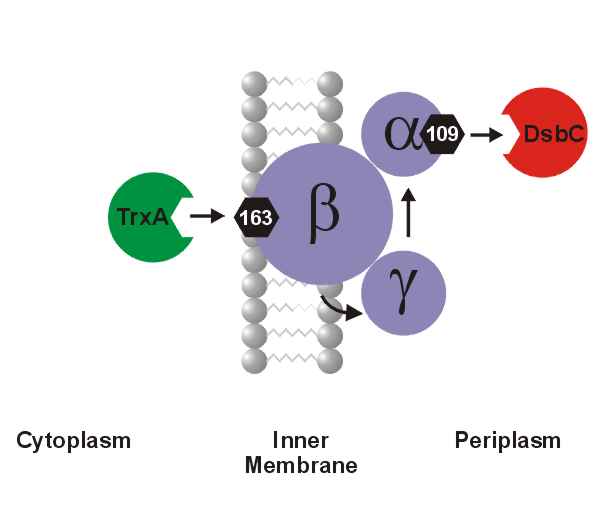
A number of secreted and membrane proteins in both bacteria and eukaryotes contain disulfide bonds. In 1991, we presented evidence that bacteria require the periplasmic protein DsbA for efficient formation of these bonds. DsbA is maintained with its active site cysteines in the oxidized state by the membrane protein DsbB. We are currently carrying out a genetic analysis of DsbA and DsbB to allow a structure-function correlation. The isomerization of incorrectly formed disulfide bonds depends on the periplasmic DsbC protein. This protein, which is maintained with its active site cysteines in the reduced state, receives its electrons from thioredoxin in the cytoplasm via a membrane protein DsbD. We are studying this interesting mechanism of electron transfer across the cytoplasmic membrane. We are also exploring more fully the role of the various disulfide reducing components of the cytoplasm. These include the thioredoxins and glutaredoxins, but we are also characterizing alternative pathways for disulfide bond reduction.
|
|
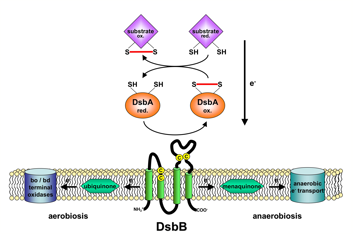 Model of the DsbA-DsbB system Model of the DsbA-DsbB system |
|
|
Oxidative and reductive disulfide branch of the E. coli periplasm |
|
|
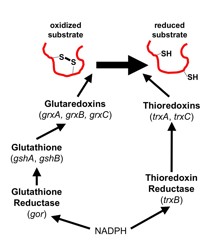
The cytoplasmic redox pathways of E. coli are responsible for the reduction of substrates such as the essential protein ribonucleotide reductase. Two pathways, the glutaredoxin and thioredoxin pathways, function in parallel, with the thioredoxins reduced by thioredoxin reductase (trxB) and the glutaredoxins reduced by glutathione, which is maintained in its reduced state by glutathione reductase (gor). Both pathways ultimately derive their reducing power from NADPH. Although this transfer of electrons down these pathways is known, many questions about the cytoplasmic redox pathways remain unanswered, such as their redundancy and specificity. Suppressor analysis has been used in the past to elucidate these pathways and discover novel biological phenomena. Suppressors of ΔtrxB Δgor are a triplet repeat expansion of ahpC to ahpC*, and in these strains an alkyl hydroxyperoxidase is converted to a disulfide reductase. Because this mutation occurs frequently, we isolated suppressors of ΔtrxB Δgor ΔahpCF in order to discover other pathways for disulfide bond reduction. In these suppressors, we have identified mutations in lpdA, the gene for lipoamide dehydrogenase. LpdA is homologous to Gor and the mutant form of this protein may perform a similar function as Gor in order to reduce disulfide bonds in these suppressors, or may be able to reduce oxidized lipoamide and transfer electrons to ribonucleotide reductase via a novel mechanism.
|
|
|
Disulfide bonds formed between pairs of cysteines are important structural features of many exported proteins. I have been studying the oxidative branch of disulfide bond formation in the periplasm. My goal is to understand the molecular events that have to occur for the in vivo formation of each disulfide bond in folding proteins.
|
|
Exploring the diversity in redox biology of the bacterial cell envelope The formation of disulfide bonds is an important event in the folding of many proteins found in the cell envelope of E. coli. The enzymes that catalyze the formation of disulfide bonds have been identified and characterized in detail in E. coli. However, there is not a great deal of information regarding this process in other bacteria. Currently, there are over 500 publicly available genome sequences, representing bacteria from diverse habitats and divergent evolutionary histories. Using a combination of bioinformatic and experimental approaches, I am interested in addressing the following questions: Is disulfide bond formation a conserved process across the Domain Bacteria? If so, are the pathways fully conserved, or are there alternative ways to make disulfide bonds? If disulfide bond formation is not conserved, which types of bacteria are missing this process? How do these bacteria compensate for a lack of disulfide bonds in protein folding pathways?
|
|
|
Understanding the specificities of E. coli glutaredoxins The cytoplasm of Escherichia coli is constantly maintained as a reducing environment in order to allow thousands of specific reactions. The cytoplasmic reduction of disulfide bonds is achieved by the thioredoxin superfamily which includes the glutaredoxin proteins along with glutathione. Members of this protein family are present in most, if not all, living organisms. Glutaredoxins catalyze reversible oxidation/reduction of protein disulfide groups and glutathione-containing mixed disulfide groups. They are involved in different cellular processes such as DNA replication, sulfate assimilation or response to oxidative stress. The variety of cellular functions performed by the glutaredoxin family is made possible, in part, by the wide range of redox potentials associated with their active site Cys-X-X-Cys motif. Glutaredoxin 1 and glutaredoxin 3 exhibit 33% sequence identity and are structurally nearly superimposable. Yet, glutaredoxin 1 can reduce the essential enzyme ribonucleotide reductase, which provides deoxyribonucleotides for DNA replication, and PAPS reductase, required for cysteine biosynthesis, while glutaredoxin 3 cannot. These two proteins provide a good model system for understanding the specificity of these proteins.
|
|
|
In Escherichia coli, the glutathione/glutaredoxin and thioredoxin pathways are essential for the reduction of cytoplasmic protein disulfide bonds, including those formed in the essential enzyme ribonucleotide reductase during its action on substrates. Double mutants lacking thioredoxin reductase (trxB) and glutathione reductase (gor) or glutathione biosynthesis (gshA) cannot grow. Growth of Δgor ΔtrxB strains is restored by a mutant (ahpC*) of the peroxiredoxin AhpC, converting it to a disulfide reductase that generates reduced glutathione. Our studies show that ahpC* also restores growth to a ΔgshB ΔtrxB strain, which lacks glutathione and accumulates only its precursor γ-glutamylcysteine (γ-Glu-Cys). Surprisingly, new ahpC suppressor mutations arose in a strain lacking both glutathione and γ-Glu-Cys, ΔgshA ΔtrxB, which ahpC* does not suppress. Our results show the plasticity of AhpC, which under selective pressure can be altered by different mutations to channel electrons into ribonucelotide reductase by at least four distinct routes, acting via glutathione, γ-Glu-Cys, the thioredoxins or glutaredoxin 1. The various suppressor strains exhibit an oxidizing cytoplasm and accumulate correctly folded alkaline phosphatase, FAB antibody or tissue plasminogen activator. Proteins most effectively oxidized vary between strains, potentially providing useful tools for expressing different disulfide-bonded proteins. |
|