|
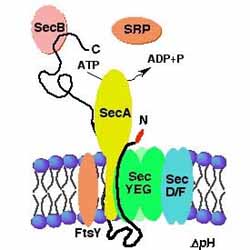
Core features of the major mechanism for the translocation of proteins across membranes are conserved throughout biology. The bacterium Escherichia coli, like all other organisms, synthesizes secreted proteins with amino-terminal signal sequences, which direct such proteins to a translocation machinery located in the cytoplasmic membrane. In this laboratory, we have used genetic approaches to identify several of the protein components of this machinery. We are currently studying the functions in this process of membrane proteins that play a key role in protein export. In particular, we are seeking mutants that alter secretion so that the transport machinery remains in the pore-open conformation. Further, we are studying the role of two proteins, SecD and SecF, which may play a role late in the translocation process. Finally, we are examining the role of features of the amino acid sequence of secreted proteins other than the signal sequence that allow such proteins to pass through the membrane. |
 |
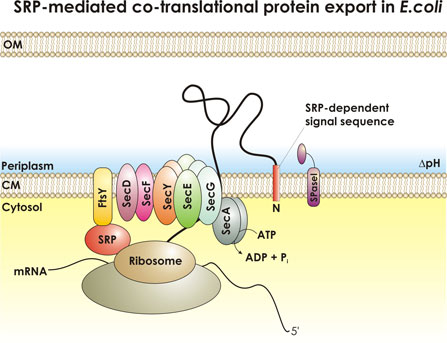
Sebastian Ahrens and Markus Eser One project in the Beckwith laboratory tries to further our understanding of protein export across the cytoplasmic membrane of E. coli. The major E. coli protein export machinery is the Sec machinery, with its core proteins SecYEG residing within the cytoplasmic membrane. Proteins that are designated to the cell envelope are synthesized in the cytoplasm as pre-proteins with N-terminal signal sequences. Signal sequences are short (about 18 amino acids) and show little sequence homology). Preproteins are recognized by parts of the export machinery and translocation is initiated either after (post-translational) or during protein synthesis (co-translational). Initiation of co-translational translocation requires the so-called signal recognition particle (SRP). Our laboratory has identified a distinct set of signal sequences that depend on the SRP, enabling us to find a trend in the characteristics of these SRP-dependent signal sequences that we think is critical for the discrimination by the SRP: SRP-dependent signal sequences generally show higher hydrophobicity than non-SRP-dependent signal sequences. I'm working on refining our model for SRP-dependent signal sequences allowing us to distinguish between SRP-dependent and non-SRP-dependent signal sequences. As a model system we are using the export of thioredoxin 1 (TrxA) to the periplasm by the Sec machinery. TrxA, natively a cytoplasmic protein can only be exported efficiently when fused to a SRP-dependent signal sequence. In our studies we found signal sequences that are very hydrophobic but do not promote the export of TrxA. I want to use these highly hydrophobic non-SRP-signal sequences to try to find mutations by random mutagenesis that allow efficient export of TrxA. We can easily isolate mutants that lead to TrxA export by a genetic selection. Once mutant signal sequences are found, we hope to broaden our knowledge on how a subset of short sequences with little sequence homology is able to efficiently designate the mode of translocation across the cytoplasmic membrane. |
|